CBPV





Chronic Bee Paralysis Virus (CBPV), is a virus that cause an infectious disease of the adult honey bees. Previously known with different names in different countries such as “little blacks” (UK), “hairless black syndrome” (US), “mal nero” (Italy), “Schwarzsucht” (Germany) and “mal noire” (France) (Ribière et al., 2008). These names reflect one of the two possible symptom sets that can be seen in CBPV infection. First, lots of honey bees appear black, hairless, shiny and greasy in bright light. Infected bees do not actively contribute to the hive tasks and usually are attacked by the normal bees by nibbling their wings (Figure 1). The second set of symptoms is characterised by inability to fly, trembling, mass paralysis and finally death (Rinderer and Rothenbuhler, 1975). The two sets of symptoms could appear in the same infected hive singularly or mixed.
Mechanisms of transmission that have been demonstrated for CBPV are injection, contact, oral (even if a large amount of virus particles are needed in this last case) and probably vertical (trough infected queens) (Bailey, 1965; Kulinevic et al., 1973; Chen et al., 2005a; Chen et al., 2006b ).
CBPV is the only honey bee virus that still cannot be assigned to any group of viruses. It displays asymmetric particles of 30-65 nm in length and about 20 nm in width. However, a plethora of shapes has been described including rings and figures of eight with dimensions up to 640 nm in length (Figure 2) (Bailey et al., 1963).
The virus is widespread in Britain , causes mortality in honey bee colonies yet CBPV does not seem to follow a seasonal pattern as seen in other viruses (Bailey et al., 1981; Tentcheva et al., 2004b). The Varroa mite is not likely to be a vector as CBPV has not been found in the parasite.
CBPV is often found in association with a ‘‘satellite’’ virus, first called Chronic paralysis virus associate (CPVA) now named Chronic bee paralysis satellite virus (CBPSV) and is classified as the only member of the Chronic bee paralysis virus associated satellite subgroup (Bailey,1976; Bailey et al., 1980; Fauquet et al., 2005). Under T.E.M. they appear as isometric particles of 17nm in diameter and they have three single-stranded RNAs of about 1100nt each and were reported to have a single capsid protein of about 15kDa (Bailey et al., 1980). CBPSV are still of unknown significance, what we know to date about them is that they are serologically unrelated to CBPV, but cannot multiply in the absence of CBPV. (Chen and Siede, 2007; Ribière et al., 2010).


Fig 1: CBPV symptoms. On the left: black bee with nibbled wings; on the right: shiny, “greasy” bees (Crown copyright, National Bee Unit, FERA).
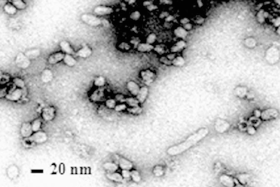
Fig. 2: CBPV (T.E.M). The virus show asymmetric particles of different dimensions. (Ribière et al., 2010).
The characteristics and the genome organization of CBPV is very different compared to the assigned viruses discussed above. CBPV has been seen to encode polypeptides of about 75, 50, 30 and 20 kDa (Olivier et al., 2008a; Ribière et al., 2000).
The RNA is composed of two separate filaments of RNA respectively called RNA1 of 3674 bp and RNA2 of 2305 bp; the 5' is capped and no poly-adenylate tail is present (Figure 3) (Oliver et al., 2008a; Ribière et al., 2010).
The ORF 3 in RNA1 shows significant similarities with the RdRp of the single-strand RNA viruses. No similarities were found in the Genbank database for the other ORFs (Ribière et al., 2010).
Matching the CBPV RdRp sequence with the sequences deposited in Genbank, using the Basic Local Alignment Search Tool (BLAST), Ribière found that this virus could have a phylogenetic position between the Nodaviridae (insect and fish viruses) and Tombusviridae (plant viruses) family clusters even if it differs from these two families in many structural and genomic characteristics (Ribière et al., 2010). The lack of similarities between this virus and other viruses described in the literature led us to consider this virus as a type species of a new group of positive stranded RNA viruses (Ribière et al., 2010). To justify this suggestion it is interesting to see the Figure 4 that report the original phylogenetic tree obtained by Ribière aligning the RdRp sequences and the reported comparative table of the characteristics of Nodaviridae, Tombusviridae and CBPV. In fact even if the RdRp shows some analogies with the RdRp of Nodaviridae and Tombusviridae, the differences in the structural characteristics (e.g. host and particle shape) don't allow CBPV to join any of the two families.
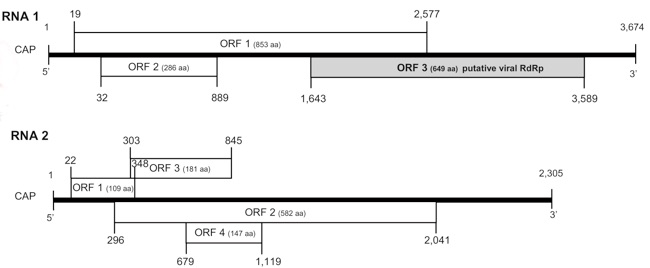
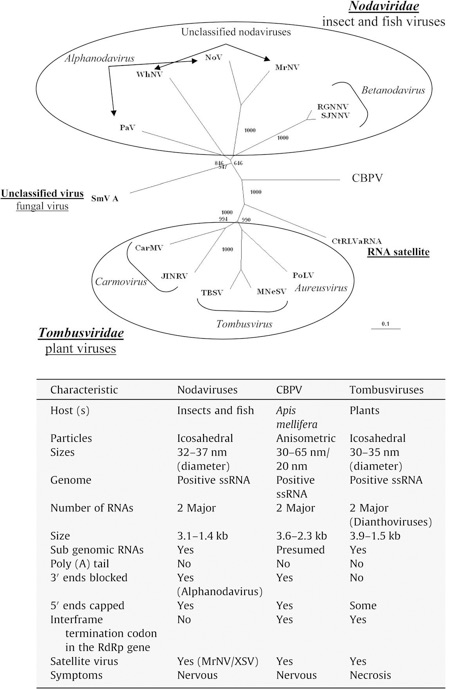
Fig 3: Scheme of the genome organization of CBPV. Predicted genome organisation of CBPV RNA 1 and RNA 2. The positions of seven putative ORFs are indicated and the putative sizes of the protein encoded as described by Ribière. Legend: CAP= capped 5', ORF 1-4= open reading frame 1-4, RdRp= RNA dependant, RNA polymerase (Ribière et al., 2010 modified)
Fig 4: Taxonomical position of CBPV. Phylogenetic tree based on RdRp sequence and a table reporting the differences between CBPV, Nodaviruses and Tombusviruses (from Ribière et al., 2010 modified)



Citation: Cordoni, G.; 2011. Epidemiology and taxonomy of honey bee viruses in England and Wales. PhD thesis. University of Surrey.
Developed by
Dr. Guido Cordoni DVM, PGDip, PhD, MRCVS