Dicistroviridae





Dicistroviridae, characteristics and genetics
The name originates from the dicistronic organization of their RNA genome. They are non-enveloped viruses showing a icosahedral T-3 symmetry. The capsid is composed of 60 repeating protomers, each consisting of a single molecule of each of three proteins VP1, VP2, and VP3. In addition to these three proteins, a smaller internal protein VP4 may also be present (e.g. BQCV and ABPV) (Govan et al,. 2000). The virion dimensions range from 17 to 30 nm. The Figure 1 shows a representation of the virion. Dicistroviruses have a monopartite, linear ssRNA(+) genome ranging from 8.5 to 10.2 kb. A viral protein (VPg) is bounded at the 5’-terminus of the viral RNA and at the 3’end there is a polyA tract (about 20 bp). The genome is organized into two non-overlapping open reading frames, ORF1 and ORF2, that encode the non-structural and structural proteins, respectively as shown in Figure 1 (Chen et al., 2007 modified).
The viral RNA is infectious and acts also as mRNA, The RNA encodes two polyproteins from ORF1 and ORF2. The first encodes the non-structural proteins involved in replication, while ORF2 encodes three (or four depending on the virus) capsid proteins. ORF1 is preceded by an untranslated region (UTR) that contains as an internal ribosome entry site (IRES) element; an Intergenic Region (IGR), upstream of the ORF2 which also contains an IRES element. At the 3' end there is another UTR upstream of the polyA tail. The length of the poly(A) tail, genetically determined, varies in different viruses and the 3' UTR, folded in a steam-loop structure, seems to be involved in RNA replication (Chen et al., 2007).
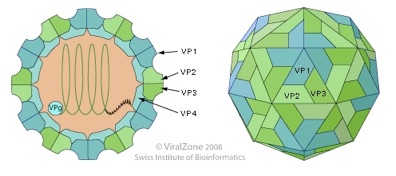
Fig.1: Representation of the virion structure. On the left picture the internal organization of the virion containing the RNA genome is shown. The capsid is formed by the four proteins VP1, VP2, VP3 and the internal VP4. On the right the classical T3 symmetry of the virion is shown; each side of the icosahedron is made by the VP1, VP2 and VP3 protein.
(http://www.expasy.org/viralzone/all_by_species/36.html (modified)).
Black queen cell virus (BQCV).
Infection of honey bees with BQCV has been reported in North America, Central America, Europe, Oceania, Asia, Africa, and the Middle East (Allen and Ball, 1996; Ellis and Munn, 2005). The virus mainly affects developing queen larvae and pupae while they are in capped cells. Diseased larvae have a pale yellow appearance and a sac-like skin, a symptom somewhat similar to that induced by Sacbrood virus (SBV), but infected pupae turn
dark in colour or black and die rapidly. The wall of the queen cell becomes dark and this is a characteristic symptom of BQCV infection. Worker bees can also be infected by BQCV but normally do not exhibit disease symptoms.
A study conducted by Tentcheva (Tentcheva et al., 2004b) indicated that BQCV infection was more prevalent in adult bees than in pupae and that the incidence of BQCV was higher in spring and summer than in autumn. BQCV seems to be transmitted through the glandular secretions of the nurse bees. Even though a positive association between Nosema and BQCV was found in the past, the mechanism by which Nosema activate and transmit BQCV infection remains to be determined (Chen and Siede, 2007).
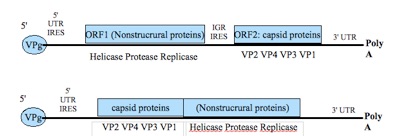
Fig.2: Genome organization of Dicistroviridae (upper) and Iflaviridae (lower):
From the left to the right: VPg= genomic viral protein; 5'UTR= untranslated region (near the 5' end); IRES= internal ribosome entry site; ORF1= open reading frame 1; IGR= intergenic region; ORF2= open reading frame 2; VP 1,2,3,4= viral protein 1,2,3,4; 3'UTR= untranslated region (near the 3' end); PolyA= poly-adenylate tail (Chen et al., 2007 modified).
The Acute bee paralysis virus (ABPV)-Kashmir bee virus (KBV)-Israeli acute paralysis virus (IAPV) complex.
Acute bee paralysis virus (ABPV), Kashmir bee virus (KBV) and Israeli acute paralysis virus (IAPV) share many characteristics, not only in their gene arrangement, but also their biological behaviour. These viruses share the same routes of transmission, (e.g. transmitted by Varroa or as a result of cannibalization of diseased larvae) and a widespread, frequently asymptomatic, distribution. Clinical symptoms are rare and associated with elevated viral titres producing symptom such as trembling, progressive paralysis, inability to fly, dark bees and loss of hair. Unlike the CBPV, the paralysis often remains at the level of the individual and rarely translates to mass paralysis (de Mirandaet al., 2010a ).
These viruses were all discovered in the laboratory while scientists were undertaking propagation experiments in white eyed pupae (Bailey et al., 1963). In fact ABPV was found in a study on CBPV, KBV in 1974 was found in a study on Apis iridescent virus in Apis cerana (Bailey et al., 1976, 1979), and IAPV was purified in 2002 while propagating the extract of a single bee from a cluster of dead bees found in front of a failing hive in Israel (from which the name of the virus is derived) (Maori et al., 2007 a,b).
The high variability in the genome of these viruses has complicated the diagnosis of disease and their classification which relies on the detection of specific viral sequence using methods based on PCR (de Miranda et al., 2010a).
IAPV, KBV and IAPV have been reported all over the world singularly and also associated together as shown in Figure 3 (de Miranda et al., 2010a ).
The presence of ABPV in A. mellifera has been reported in North America, Central and South America, Europe, Oceania, Asia, Africa, and the Middle East (Ellis and Munn, 2005). Infection with ABPV rarely causes clinical signs (Bailey et al., 1981) and is thus often seen in apparently healthy adult bees, particularly during the summer. Spread of ABPV in the colonies is probably via salivary gland secretion of infected adult bees. (Chen and Siede, 2007). ABPV is considered to be the second most-prevalent virus of the honey bee in Austria (Berenyi et al., 2006).
Varroa mites can act as a virus vector and transmit ABPV. Detection of large amounts of the virus in diseased or dead bees from colonies heavily infested with Varroa mites suggests that infestation of Varroa mites may also stimulate the virus to replicate and favour disease induction over asymptomatic infection (Chen and Siede, 2007). While Varroa mites might activate ABPV, inducing depletion in the defence of the bees and directly infecting virus into the haemolymph, the replication of the virus can be also induced by other factors. Some studies showed that ABPV was present in bees from apiaries where no APBV-positive Varroa mites were detected (Tentcheva et al., 2004b) and that replication of ABPV can be activated by simple stress procedures such as injection of potassium phosphate buffer (Hung et al., 1996). This suggests that the Varroa mite is not the sole factor contributing to outbreaks of ABPV infection (Chen and Siede, 2007).
Strains of KBV have been found in A. mellifera from Canada (Allen and Ball, 1995), Fiji (Anderson, 1991), Spain (Allen and Ball, 1995), and the United States (Bruce et al., 1995; Hung et al., 1995). KBV attacks all stages of the bee life cycle and persists within the brood and adult bees as an unapparent infection (Ball, 1985; Anderson and Gibbs; 1988). The disease and associated mortality lack clearly defined symptoms. (Chen and Siede, 2007). KBV is considered to be the most virulent honey bee virus under laboratory conditions. It multiplies quickly if introduced into the haemolymph, causing mortality within 3 days. It has been shown that Varroa mites are effective vectors of KBV passing this virus by injecting it into the host haemolymph (Chen et al., 2004). KBV seems not to be infectious through food (Chen and Siede, 2007).
Immuno-diffusion tests, using the structural proteins as antigen in the production of antibodies, showed that strains of KBV from Canada and Spain are serologically more closely related to ABPV than to other KBV strains (Allen and Ball, 1995). Molecular analysis revealed KBV and ABPV share about 70% sequence homology over the entire genome, although there are significant
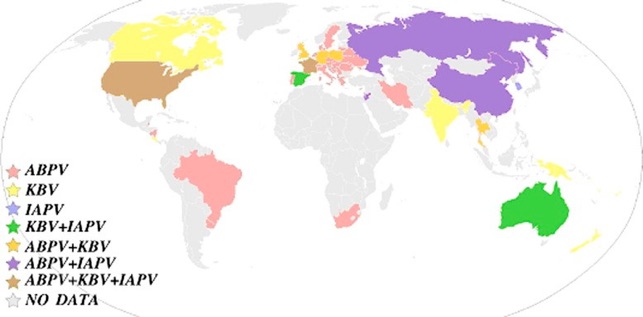
Fig.3: World distribution of ABPV, KBV and IAPV by country. Data were compiled from Allen and Ball (1996), Ellis and Munn (2005), recent publications and from published and unpublished GenBank database entries. The colour codes for the presence of single or multiple viruses are shown. (de Miranda et al., 2010a ).
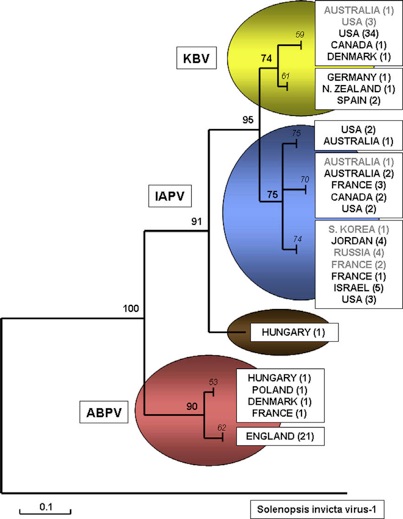
Fig 4: Relationships between KBV, IAPV and ABPV isolates. A 349 nt section within the polymerase region (nt 5454–5802 of the KBV genome) was aligned, using Solenopsis invicta virus-1 (SnIV-1) as an outgroup. The phylogram was constructed by MEGA-4 (Tamura et al., 2007), using Minimum Evolution criteria and 1000 replicate bootstrap analysis; branches with less than 60% bootstrap support were collapsed.
Legend: bold = internal branch; italic = terminal node leading to each taxonomic genogroup; grey type= IAPV/ KBV isolates currently misclassified in the public DNA databases;black type = isolates correctly classified. (de Miranda et al., 2010a).
differences in several critical areas of the genomes between the two viruses (de Miranda et al., 2010a ). KBV infection is less prevalent than the other bee viruses. A large-scale field survey in France showed that KBV was found in the adult population in only 17% of the apiaries (Tentcheva et al., 2004b) but this data could be affected by miss-classification between KBV and IAPV that was actually found in France recently (Blanchard et al., 2008b).
IAPV is the most recently identified honey bee virus of this group and it is surprisingly very closely related to KBV genetically (Maori et al., 2007a) as it is possible to see in the figure 4 that shows a phylogenetic tree of KBV/ABPV/IAPV isolates using Solenopsis Invicta virus 1 as outgroup. This relationship is sufficiently close that IAPV may be better regarded as a variant of established viruses (KBV) rather than as a distinct virus in its own right. It is considered by some authors (Cox-Foster et al., 2007) as a marker for Colony Collapse Disorder (CCD), the syndrome that is jeopardising honey bees in America. It was considered a marker because he was found in all the hives with CCD (but it was also found in healthy hives). However, further studies are required to clarify this role (Anderson and East, 2008).



Citation: Cordoni, G.; 2011. Epidemiology and taxonomy of honey bee viruses in England and Wales. PhD thesis. University of Surrey.
Developed by
Dr. Guido Cordoni DVM, PGDip, PhD, MRCVS